Saito, M. A., Saunders, J. K., McIlvin, M. R., Bertrand, E. M., Breier, J. A., Brisbin, M. M., Colston, S. M., Compton, J. R., Griffin, T. J., Hervey, W. J., Hettich, R. L., Jagtap, P. D., Janech, M., Johnson, R., Keil, R., Kleikamp, H., Leary, D., Martens, L., McCain, J. S. P., Moore, E., Mehta, S., Moran, D. M., Neibauer, J., Neely, B. A., Jakuba, M. V., Johnson, J., Duffy, M., Herndl, G. J., Giannone, R., Mueller, R., Nunn, B. L., Pabst, M., Peters, S., Rajczewski, A., Rowland, E., Searle, B., Van Den Bossche, T., Vora, G. J., Waldbauer, J. R., Zheng, H., and Zhao, Z.: Results from a multi-laboratory ocean metaproteomic intercomparison: effects of LC-MS acquisition and data analysis procedures, Biogeosciences, 21, 4889–4908, https://doi.org/10.5194/bg-21-4889-2024, 2024.
Objective: Ocean metaproteomics is an exciting new datatype that has the potential to provide valuable new insights into the metabolic functions of marine microbes and their impact on ecological and biogeochemical processes. However, as for most new measurement types there are uncertainties associated with the accuracy and precision of measurements due to the limited extent of the application of analyses thus far, and hence there is a need to generate community confidence in metaproteomics. We propose to initiate an intercomparison effort whereby an ocean metaproteome sample from the Bermuda Atlantic Time Series is collected, divided and shared among multiple laboratories for global and targeted metaproteomic analyses. The results will be collated and discussed at a workshop of intercomparison participants. In addition, an informatic intercomparison will be conducted using a representative mass spectra data file. This effort is a follow up of the 2010 OCB scoping workshop “ The Molecular Biology of Biogeochemistry: Using molecular methods to link ocean chemistry with biological activity" and NSF EarthCube workshop that assembled US and Canadian scientists involved in metaproteomic research in May of 2017: Ocean Proteomics Data Sharing and Best Practices Workshop (Report).
Saito et al. (2019). Progress and Challenges in Ocean Metaproteomics and Proposed Best Practices for Data Sharing. J. Proteome Res. 18(4), 1461-1476.
PIs: Mak Saito and Matthew McIlvin (Woods Hole Oceanographic Institution)
September 16-17, 2021: Ocean Metaproteomics Intercomparison Workshop (virtual, Zoom)


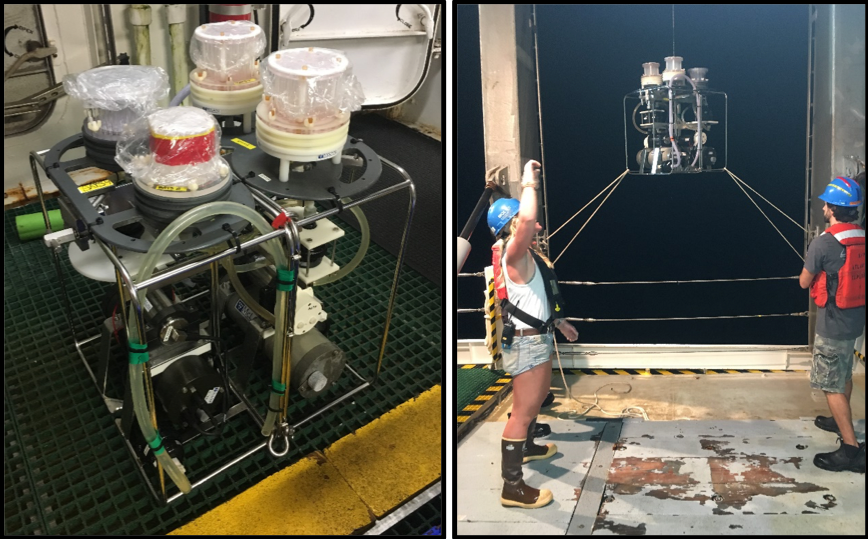
Two McLane pump samplers with two Mini-MULVS sampler heads clamped together and deployed on BATS expedition 348 on June 16th 2018 aboard the R/V Atlantic Explorer.
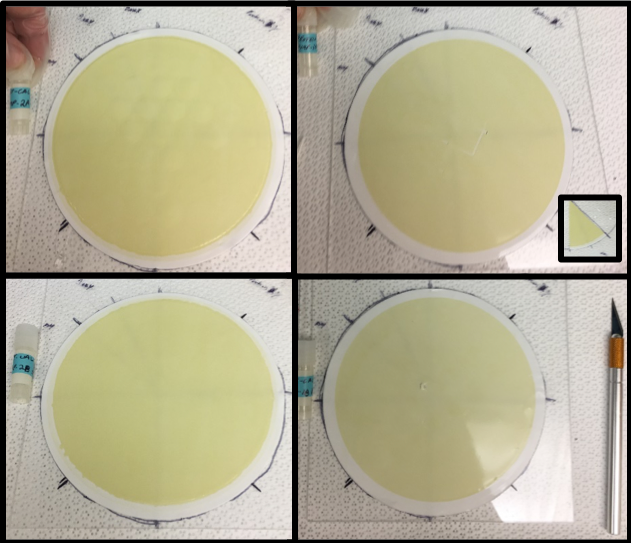
The four 142 mm filter 0.2 μm filters used for this intercomparison study collected by McLane pump (X-Acto knife for scale). Each filter was sliced into 8 fractions (inset) and frozen at -80C in a cryovial. Samples were labeled by pump and pump head (Table 1; pump-2A upper left; pump-1A upper right; pump-2B lower left; pump-1B lower right).
Acknowledgments
Funds for distributing intercomparison samples and a future workshop to discuss results is being supported by Ocean Carbon and Biogeochemistry (OCB). Funds for the research expedition where samples were collected were supported by the US National Science Foundation Biological Oceanography and Chemical Oceanography and the Gordon and Betty Moore Foundation. We thank Sophie Colston, Judson Harvey, and Gary Vora (Naval Research Laboratory) for metagenomic sequencing and Jaci Saunders (WHOI) for assembly and annotation of the paired sample.



